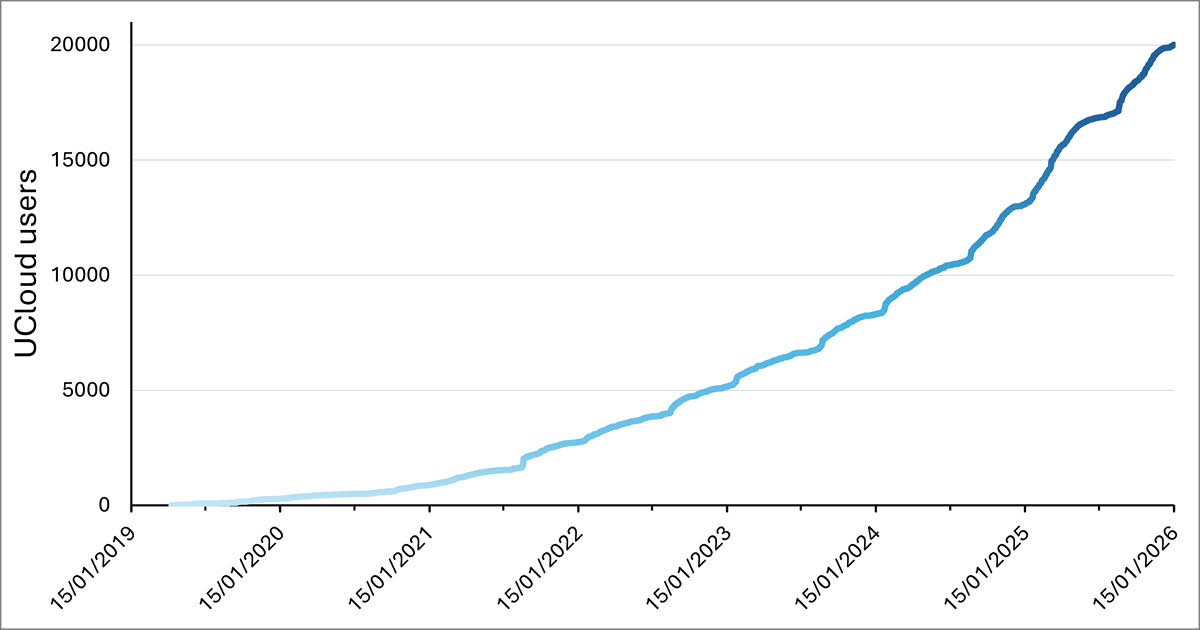
In the Master’s programme in Cognitive Science at Aarhus University, UCloud plays a central role in teaching Natural Language Processing (NLP). For instructor and PhD student Mina Almasi, the platform is essential in enabling students to work hands-on with complex models – regardless of the limitations of their own computers.
From Theory to Hands-On Learning
In a white classroom in Nobelparken, Mina stands in front of 15 students. On the screen behind her, lines of Python code appear in neat, symmetrical rows as she explains which code libraries the students need to access.

In her teaching, she uses the Coder Python application in UCloud because the course is based on Python programming. But the choice of platform is not just about software – it is about giving students the opportunity to translate theory into practice.
According to Mina, NLP teaching previously tended to remain at a more theoretical level, due to limited access to both models and the computing power needed to test theories in practice – especially when it came to large language models. With UCloud, students can now work directly with language models (LLMs) and make use of powerful GPUs and CPUs. This allows them to test theories themselves and experiment hands-on with the tools they are learning about.
“We still teach the theory, but now we can also have students use the tools in practice. They can code on their own and gain insight into how a large language model works by working directly with it through UCloud,” she explains.
A Standardised Setup that Democratises the Classroom
Another advantage of using UCloud in NLP teaching is that the platform ensures equal access for all students, regardless of the computer they own.
“There is a kind of democratisation of the classroom, because you don’t need the latest computer. You can use a five-year-old machine to run very heavy tasks that the newest tools in Natural Language Processing require,” she explains.
At the same time, the standardised setup makes teaching more seamless. All students work with the same standard configuration in UCloud, so any issues that arise are the same for everyone. This creates a shared sense of problem-solving, as challenges can be addressed collectively rather than handled individually by students on their own. As Mina puts it:
“Instead of stopping the lesson to solve individual problems, the problems become collective and an opportunity for learning for everyone. If we have a software issue – for example, a Python library version that is outdated or incompatible – it affects everyone, and we can solve it together.”
Preparing Students for Working Life
For Mina, using UCloud also helps prepare students for the reality that awaits them after graduation. According to her, many of the students who go on to IT positions will likely use cloud computing platforms rather than coding on local machines. In this way, the teaching becomes direct preparation for future job tasks and gives students experience with the technologies they will encounter in practice.

Advice for Other Instructors
Mina has used UCloud since her bachelor’s degree and finds that the platform makes teaching both smoother and more engaging.
“I recommend that other instructors make use of the platform. You just have to get started – but feel free to ask colleagues for advice on how they use it. Get some inspiration, because UCloud is a fantastic tool. It can do a great many things, but like other systems, it can feel a bit overwhelming at first, so it’s a good idea to get some guidance along the way before you begin.”